The goals of the aba package are as follows:
- to facilitate sound statistical modelling in clinical research
- to provide advanced tools for biomarker-based planning of clinical trials
- to generate publication-ready tables and figures
overview image
With the aba package, it becomes easier to fit statistical models on permuatations of covariates / predictors / outcomes / groups, to make it easier to investigate how biomarkers can be used to plan (screening / enirichment) and analyze clinical trials, and to facilitate multi-cohort validation studies when data cannot be easily shared between collaborators.
Getting started
To view tutorials and examples of how to use the package, you can visit the package website at https://ncullen93.github.io/abaR/.
You can install the development version of the aba package from GitHub with the following:
# install.packages("devtools")
devtools::install_github("ncullen93/abaR")The typical workflow for performing statistical analysis with aba is incredibly simple:
library(aba)
df <- aba::adnimerge %>% filter(VISCODE == 'bl')
# build and fit an aba model with multiple outcomes and predictors
model <- df %>% aba_model() %>%
set_outcomes(ConvertedToAlzheimers, CSF_ABETA_STATUS_bl) %>%
set_predictors(
PLASMA_ABETA_bl, PLASMA_PTAU181_bl, PLASMA_NFL_bl,
c(PLASMA_ABETA_bl, PLASMA_PTAU181_bl, PLASMA_NFL_bl)
) %>%
set_stats('glm') %>%
fit()
# summarise the model results (coefficients and metrics)
model_summary <- model_fit %>% summary()You can then generate publication-ready figures in just one line of code:
fig <- model_summary %>% aba_plot_coef(palette = 'lancet')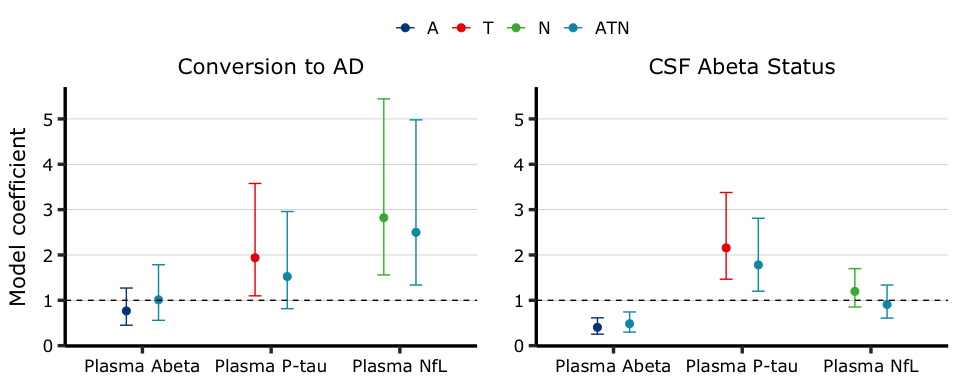
coefficient image
Publications
Some publications whose analysis has been done partially or completely with functions from the aba package are featured here:
Individualized prognosis of cognitive decline and dementia in mild cognitive impairment based on plasma biomarker combinations NC Cullen, A Leuzy, S Palmqvist, S Janelidze, E Stomrud, P Pesini, et al. Nature Aging, 2021
Prediction of future Alzheimer’s disease dementia using plasma phospho-tau combined with other accessible measures S Palmqvist, P Tideman, N Cullen, H Zetterberg, K Blennow, JL Dage, et al. Nature Medicine, 2021
Plasma biomarkers of Alzheimer’s disease improve prediction of cognitive decline in cognitively unimpaired elderly populations NC Cullen, A Leuzy, S Janelidze, S Palmqvist, AL Svenningsson, et al. Nature Communications, 2021
Comparing progression biomarkers in clinical trials of early Alzheimer’s disease NC Cullen, H Zetterberg, PS Insel, B Olsson, U Andreasson, et al. Annals of Clinical and Translational Neurology 7 (9), 1661-1673
Cite
If you use the aba package in your work, we appreciate if you cite us. An example statement which can be added at the end of the “Statistical analysis” section of your methods is provided here:
The analysis was done in the R programming language (v4.0.3) using the Automated Biomarker Analysis for R (“aba”) package [citation].